In their latest work, the AutoLC team of the CAST group of Pirok at the University of Amsterdam extended their method-development workflow to facilitate optimisation of 2D-LC separations [1].
The demonstrated workflow was capable of unsupervised gradient optimization for comprehensive 2D-LC-MS methods without needing to specify sample information. The required algorithms were designed by CAST researchers Stef Molenaar, Tijmen Bos and Jim Boelrijk, under the supervision of Bob Pirok.
The workflow was inspired by the original theoretical paper by Pirok in 2016 [2], in which retention models are constructed to computationally simulate methods and select the optimal parameters. In a nutshell, the system first utilizes very generic methods to measure the sample. This data is then used to construct retention models that describe the retention behavior of the analytes as a function of the mobile-phase composition. A large number of methods are then simulated computationally, and a computed optimum is submitted to the LC system. The LC system then carries out the proposed method and submits its data back to the system afterwards. Consequently, unsupervised optimisation of 2D-LC separations may be attained.
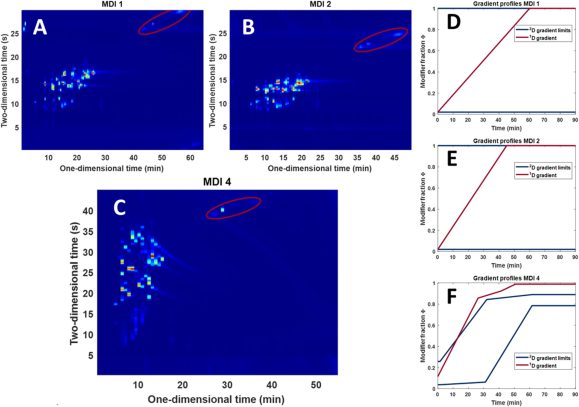
The Pirok group earlier had demonstrated their AutoLC platform on 1D separations [3], but now extended this to the more complicated 2D separation methods [1]. To accomplish this, algorithms were designed to compute and optimize complex second-dimension shifted-gradient assemblies.
The researchers also investigated the robustness of the retention models and the influence of the errors in peak-width predictions.
The study was conducted in a collaboration with the group of Dwight Stoll at Gustavus Adolphus College where the measurements were conducted. The developed tool for optimisation of 2D-LC separations was demonstrated on a complex peptide digest sample with RPLC in both of the dimensions.
The work was a product of Pirok’s UPSTAIRS (Unleashing the Potential of Separation Technology to Achieve Innovation in Research and Society) project, which is financed by the Dutch Research Council (NWO), as well as the synergetic UNMATCHED project, which is supported by BASF, DSM and Nouryon, and also receives funding from NWO.
The work was published open-access in Journal of Chromatography A and can be accessed free of charge here.
References
- Computer-driven optimization of complex gradients in comprehensive two-dimensional liquid chromatography, S.R.A. Molenaar, T.S. Bos, J. Boelrijk, T.A. Dahlseid, D.R. Stoll, B.W.J. Pirok, Chromatogr. A, 2023, 1707, 464396, DOI: 10.1016/j.chroma.2023.464306.
- Program for the interpretive optimization of two-dimensional resolution, B.W.J. Pirok, S. Pous-Torres, C. Ortiz-Bolsico, G. Vivó-Truyols and P.J. Schoenmakers, J. Chromatogr. A, 2016, 1450, 29–37, DOI: 10.1016/j.chroma.2016.04.061.
- Chemometric Strategies for Fully Automated Interpretive Method Development in Liquid Chromatography S. Bos, J. Boelrijk, S.R.A. Molenaar, B. van ‘t Veer, L.E. Niezen, D. van Herwerden, S. Samanipour, D.R. Stoll, P. Forré, B. Ensing, G.W. Somsen, B.W.J. Pirok, Anal. Chem. 2022, 94(46), 16060–16068, DOI: 10.1021/acs.analchem.2c03160.

